Notable changes in Auspice
The Nextstrain team
The Nextstrain team continues to improve the visualisation capabilities of Auspice, including new features, bug fixes and performance improvements. This year, we've released 19 versions and wanted to highlight some of the significant changes.
# Table of contents
- View in other platforms
- Performance improvements
- Convey tip confidence through saturation
- Focus on selected
- Filter on all metadata
- Full changelog
# View in other platforms
Historically, we have always been strong advocates for interoperability between platforms. For many years, Nextstrain.org has supported fetching HTTPS-addressable Auspice JSONs. UShER uses this feature to link to Auspice views for subtrees.
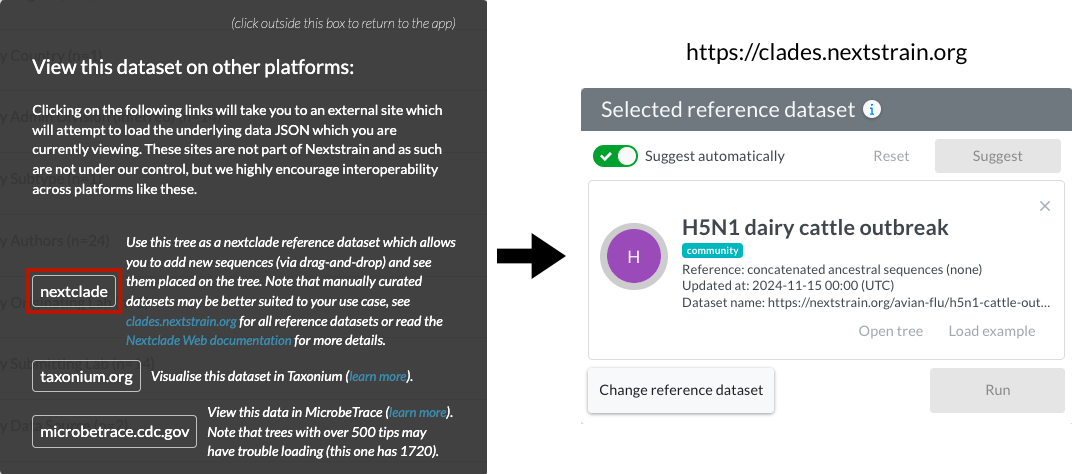
Other platforms have developed a similar approach to access Auspice datasets, often using our RESTful API. When viewing datasets in Nextstrain, you can switch to supported platforms via the new VIEW IN OTHER PLATFORMS button at the bottom of the page:
There are currently three supported platforms:
Taxonium by Theo Sanderson (added in v2.53) – allows phylogenetic visualization of large datasets.
MicrobeTrace by US CDC (added in v2.53) – allows network visualization of the Auspice phylogeny.
Nextclade (added in v2.54) – allows you to add your own sequences (in-browser) to the current tree. This feature is particularly useful to view additional sequences on top of the latest Nextstrain view of circulating diversity. Certain requirements must be met for this to work as expected, such as the root-sequence being defined for the dataset. You may get more reliable results using the manually curated reference Nextclade datasets, so please double check results against those.
# Performance improvements
As sequencing becomes more routine, the number of samples used in trees has also increased. Auspice tends to slow down for large trees (drawing performance scales with the number of nodes), diverse genomes (calculating entropy scales with the number of observed mutations) and diverse metadata and geographic spread (the map complexity scales with number of color-by categories and number of locations).
We've made small steps towards improving performance in Auspice. Specifically, entropy calculations are now skipped when the panel is off-screen or not rendered, and animations are now disabled for operations on trees with over 4,000 tips. These changes should result in a more responsive feel, especially on large datasets.
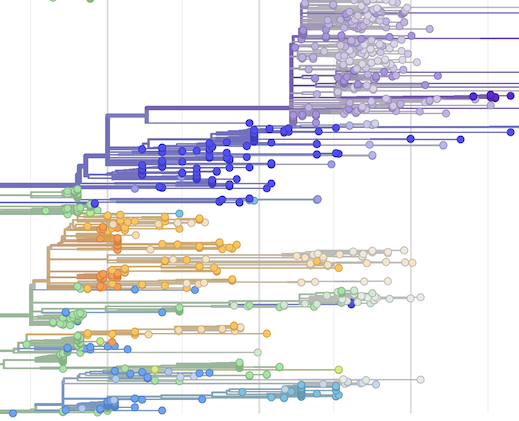
# Convey tip confidence through saturation
We've long used desaturation to convey (lack of) confidence in inferred metadata for branches, and we now do the same for tips. Tips with high confidence will still appear the same as in previous versions of Auspice. We are actively using this in the H5N1 cattle-outbreak dataset where many tips lack geographic metadata:
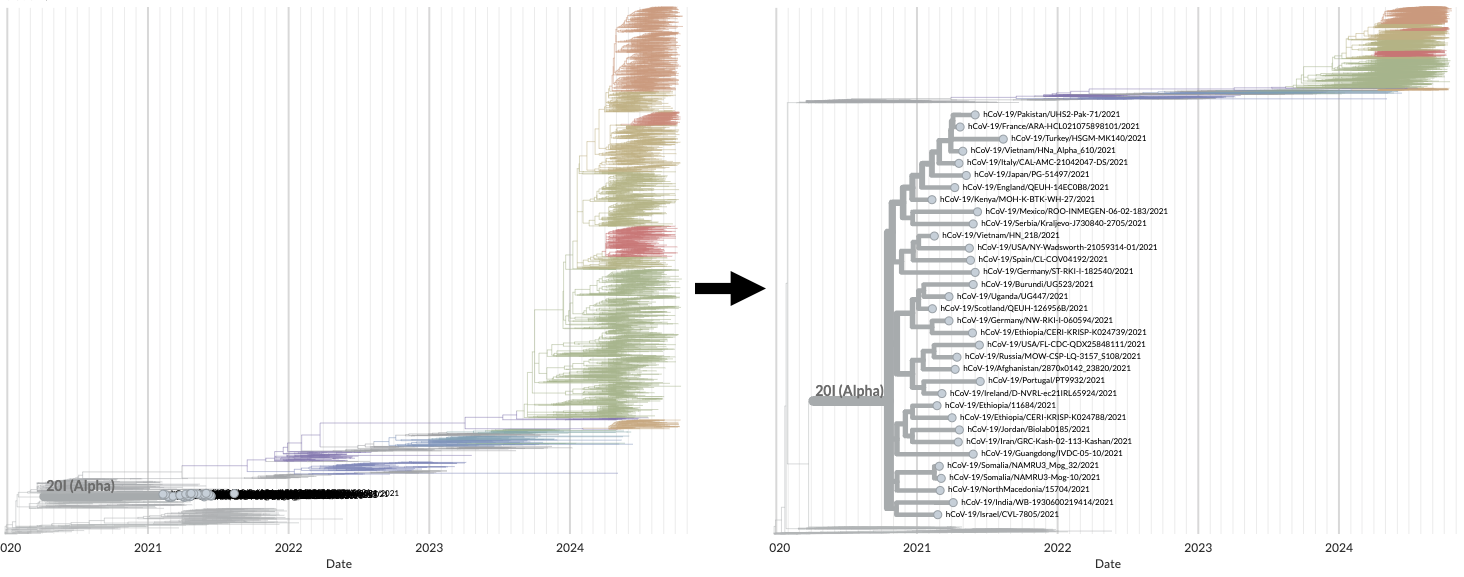
# Focus on selected
When viewing large phylogenetic trees with a sparse filter, it can be difficult to really see and understand the selected tips as they occupy a minority of the vertical space available. Enabling Focus on Selected allows the selected tips to take up more vertical space compared to other tips. For example, filtering the SARS-CoV-2 global 6m view to one of the earliest clades, 20I (Alpha):
You can toggle this focus on and off in the sidebar:
We plan to continue exploring better ways to understand sets of sequences in the context of the larger tree, initially by better use of the horizontal axis which should help for early clades in the tree such as 20I (Alpha).
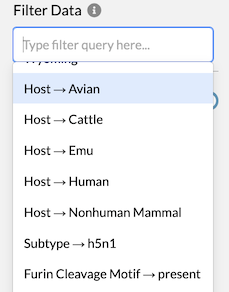
# Filter on all metadata
All categorical metadata available for the tree – i.e. all data within
node.node_attrs – are now available as options for Filter Data. Combined
with the Focus on Selected toggle described above,
this provides a powerful way to explore subsets of data.
# Full changelog
Please see the changelog for a complete summary of all changes.