New Resources for Measles Virus
Kim Andrews, Jover Lee, James Hadfield, Jennifer Chang, Trevor Bedford
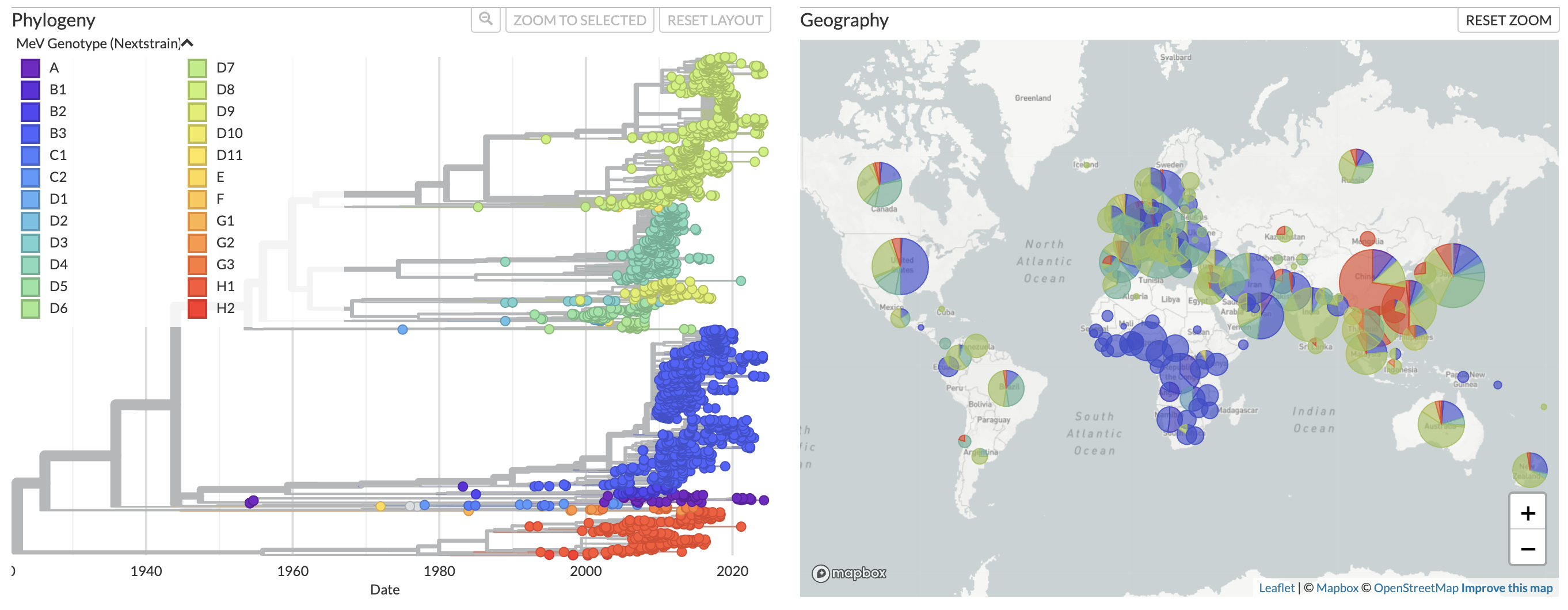
We now provide regularly updated phylogenetic monitoring of measles virus at nextstrain.org/measles. This site displays phylogenies generated using genomic data from NCBI GenBank, and is updated daily when new sequences are uploaded to NCBI. You can choose to display one of two different phylogenies based on the following data types:
- Full genome sequences
- 450bp region of the N gene ("N450")
In both phylogenies, sequences are classified to genotype using the nomenclature system established by the World Health Organization.
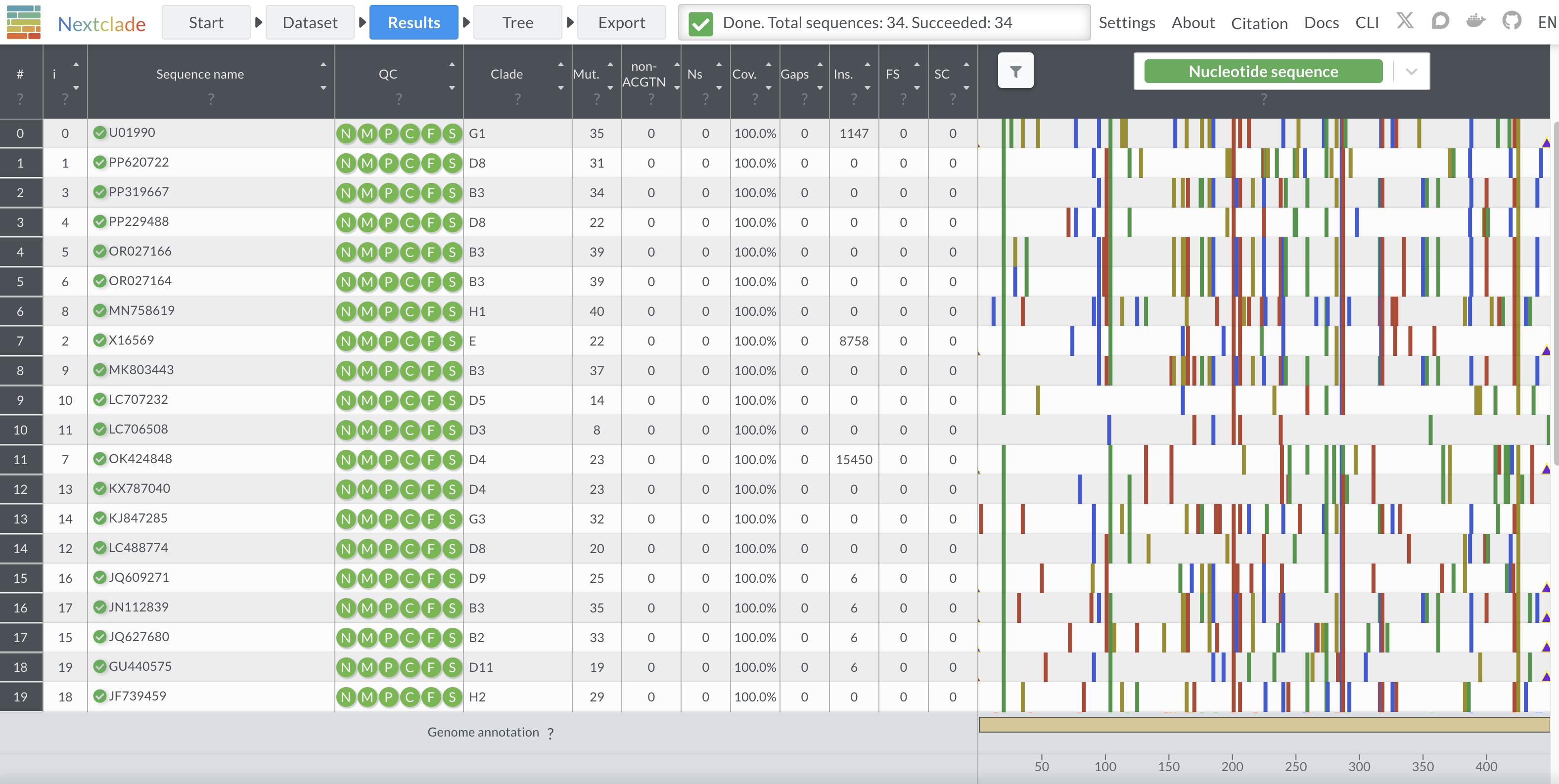
As part of this work, we developed an open-source tool for classifying measles genotypes. This tool is a Nextclade dataset, which allows users to drag-and-drop sequences onto a web browser to obtain genotype classifications. It also outputs sequence quality control metrics and displays user sequences on a phylogenetic tree.
We also provide links, updated daily, where all NCBI GenBank measles sequences and metadata can be downloaded. These currently include over 23,000 sequences, of which ~1,000 are full genome sequences, and ~19,000 include the N450 region.
With measles cases currently increasing globally, we expect these tools to be useful resources for tracking the spread of this viral pathogen. This work is made possible by the open sharing of genetic data by research groups from all over the world. We gratefully acknowledge their contributions.
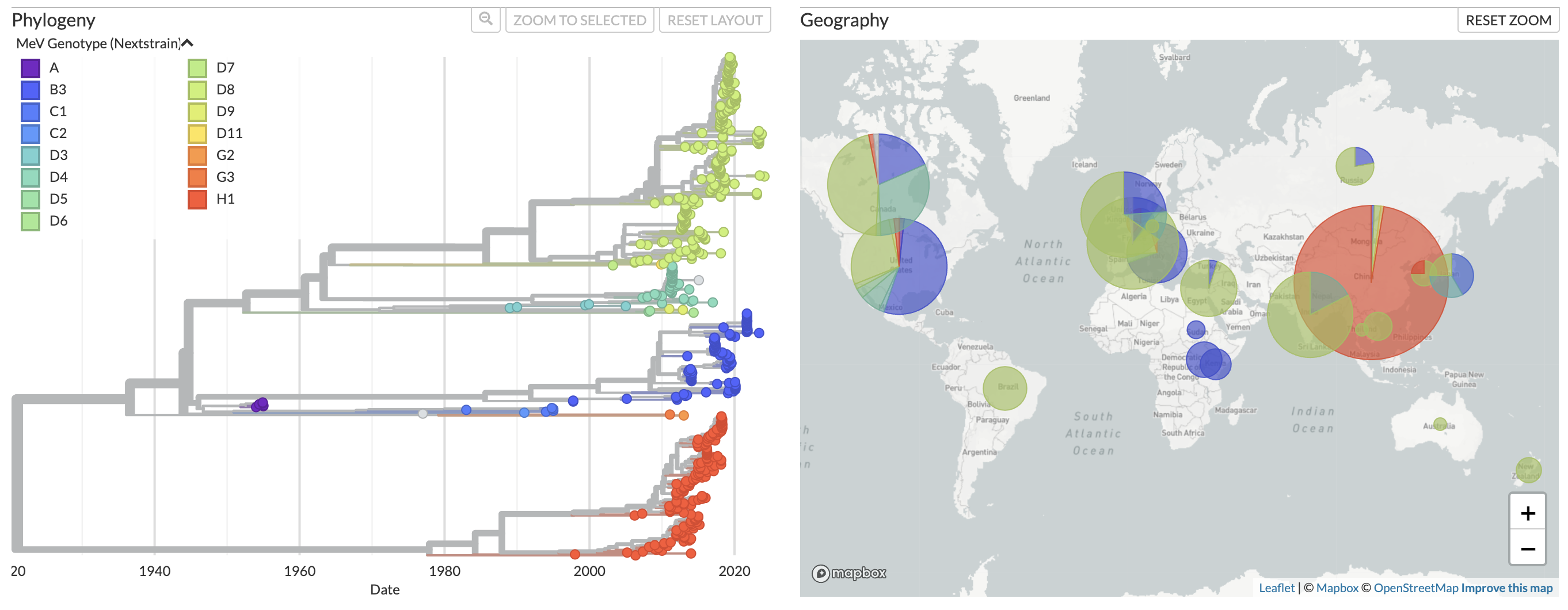 Fig 1. Phylogeny of full genome sequences for measles.
Fig 1. Phylogeny of full genome sequences for measles.