Auspice v2 released
James Hadfield
# What's New
- A new JSON format for datasets, which condenses metadata and tree data into a single JSON and allows for expanded functionality in v2.
- A whole host of UI updates to improve Auspice's visualisations, including pie charts, better rendering of dates, and more easily accessible metadata.
Read on for rationale behind this update and a detailed explanation of the changes.
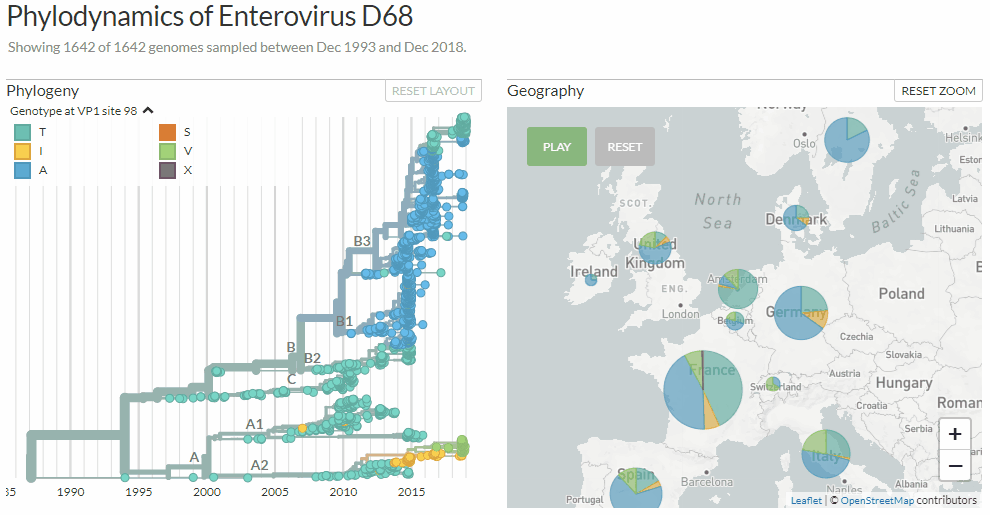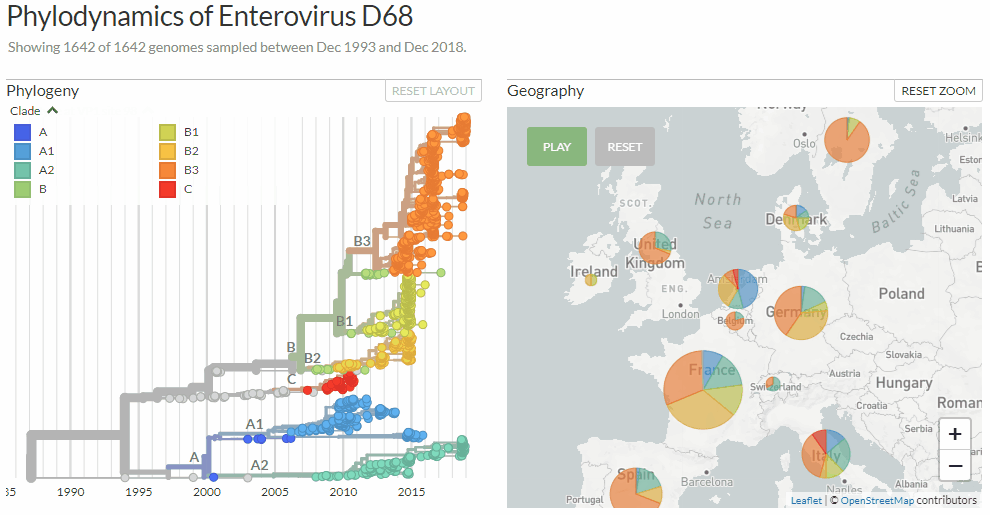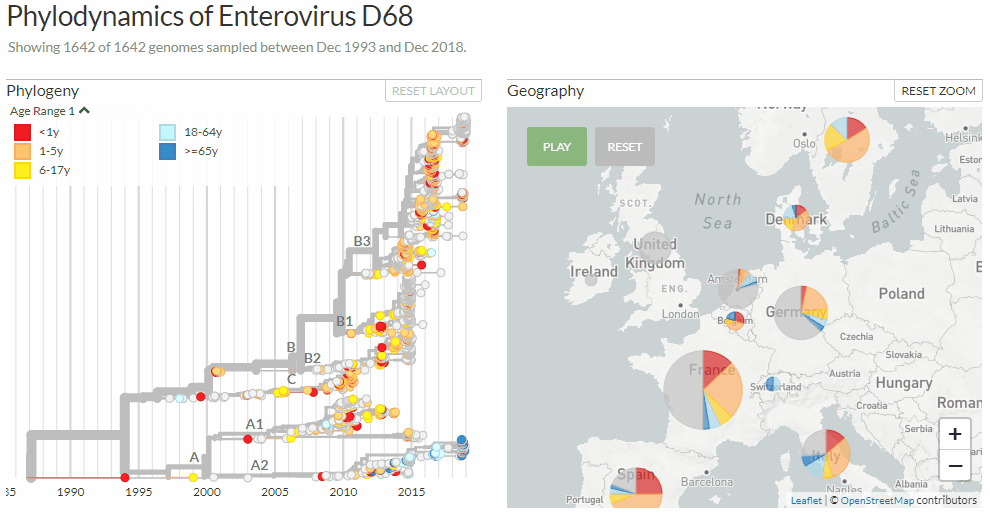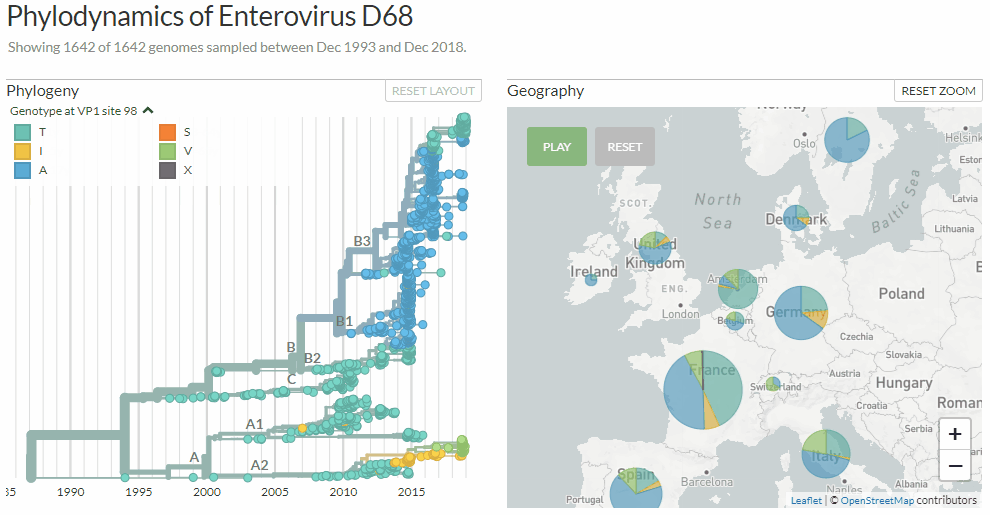 Switching between colorings in Auspice v2
Switching between colorings in Auspice v2
# Backstory
Auspice started as the nextstrain.org visualisation tool, in fact for a long time nextstrain.org was Auspice. Slowly, we turned Auspice into a stand-alone tool and started expanding its capacity. This resulted in many v1.x releases (we made it all the way to v1.39.0), and things had a habit of changing a little too frequently, with not enough focus on documentation and communication.
So in a way, this is the first proper Auspice release! There are a few big and visible changes, including using pie charts to represent discrete variables on a map, but a lot of the changes were done to make things easier behind the scenes. There are also a bunch of really cool features such as the ability to define custom servers, change the aesthetics of the Auspice client, display multiple trees and write narratives which were technically part of Auspice v1 but undocumented. This documentation website is now up to date and should allow you to understand how to use Auspice to its full potential (contributions are always welcome!)
In general, we hope that this documentation and the help messages of the various auspice subcommands are vastly improved.
The major changes which Auspice v2 brings are detailed below 👇
If you have any comments please email us or say hi on twitter.
# I don't want to upgrade - how can I continue to use Auspice v1?
While we don't think there is any reason not to upgrade, version 1 will always be available via npm:
npm install --global auspice@version1
# Pie charts to represent discrete variables on a map
We used to use colour blending to represent all the different traits present at a given location. This worked pretty well for continuous traits (e.g. the dates of isolates in each country), but performed poorly for discrete traits (e.g. the different flu clades present in each country). We now use pie charts for discrete traits which give a much nicer summary of the data:
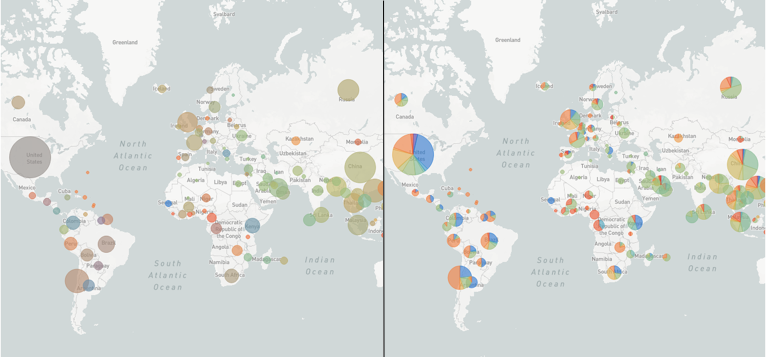 Notice the difference? Auspice v1 (left) & v2 (right)
Notice the difference? Auspice v1 (left) & v2 (right)
# New dataset JSON format
For about a year we've been seeing the limitations of the "v1" meta + tree JSONs (the dataset files behind Auspice). Additionally the format of the input JSONs to Auspice changed a bit throughout the lifetime of Auspice v1 - fine when it's an internal tool, but not so great when other people start to use it!
We've settled on a new single "v2" JSON, containing pretty similar information to the v1 JSONs but in a format which allows us to expand the functionality of Auspice.
Don't panic - auspice view and nextstrain.org will automatically convert the "v1" JSONs into the new format for you so all of the old datasets will continue to work with Auspice v2.
| file | schema | auspice versions | description |
|---|---|---|---|
| "tree" JSON | Link | v1, v2 | Decorated phylogenetic tree |
| "meta" JSON | Link | v1, v2 | The "metadata" associated with a phylogenetic tree |
| "v2" JSON | Link | v2 | The single input format required for Auspice v2 |
If you use Augur to construct your JSONs, then the next version of Augur (v6) will give you the option to produce "v1" or "v2" JSONs. If you have specific queries, the JSON schemas (above) should help you out, but here is a list of the main changes (with links to PRs where relevant, if you're really keen!):
# Strings parsed unchanged
For historical reasons, we used to "prettify" certain strings in Auspice - e.g. "usa" became "USA", "new_zealand" became "New Zealand". We dug ourselves into a bit of a hole here, as you can imagine the list of exceptions kept growing and growing. Auspice v2 now generally parses strings as they appear in the JSON, allowing you full control over how things appear. (For backwards compatability, we've kept the "prettifying" function in the v1-v2 conversion logic so the look of v1 JSONs shouldn't change.)
# Both metadata and tree data in a single JSON
The most visible change is probably the reduction of two JSONs (_meta.json + _tree.json) into a single JSON.
This single JSON has three required (and no optional) keys:
{
"version": "schema version",
"meta": {},
"tree": {}
}
# Gene / Genome definitions are now in GFF format.
v1 Auspice JSONs used 0-based starts for the gene positions and 1/-1 for which strand the gene was on.
We now use one-based start & end coordinates, following GFF format, and a "+" or "-" to denote the strand.
These definitions are now under the genome_annotations property (formerly annotations).
See PR #770.
# Changes to how node data is stored
Each tree node (internal & terminal) can now contain the following properties
name(required) -- formerly this wasstrainnode_attrs-- attributes associated with the node (sequence, date, location) as opposed to changes from one node to another.- Node attributes can now be objects and contain confidence information if available.
- A
hiddennode attribute can control Auspice's display of the node - Author information is now contained under the
authorkey, and theauthor_infodictionary is no longer present in the JSON.
branch_attrs-- attributes associated with the branch from the parent node to this node, such as branch lengths, mutations, support valuesbranch_attrs.mutations-- both AA & nucleotide mutations are now defined in the same object.
children(unchanged)
# Colorings, geographic resolutions, and defaults
The colorings property (formerly color_options) is now an array of objects, the properties of which are easier to understand.
This guarantees the ordering appears in Auspice as you define it in the JSON.
See PR #748.
The geo_resolutions property (formerly geo) is similarly an array of objects.
The display_defaults property (formerly defaults) now contains keys which are snake_case instead of camelCase.
# Multiple maintainers
The maintainer, displayed in the Auspice footer, was previously limited to a single string value and corresponding URL. We now allow multiple maintainers, each with their own (optional) URL.
# Continous, Categorical, Ordinal, and Boolean Color Scales
Traits with the "boolean" colour type which will use a pre-defined yellow & blue colour scale. Currently "continous" and "categorical" scales both use the same colour scale. Note that "discrete" types from v1 JSONs will be interpreted as "categorical".
# More information in tree info boxes
We've made more things available when you hover over the tree or click on a tree tip. For instance, v1 would use the aa-nt toggle in the entropy panel to decide which mutations to display, and it was frustrating to have to scroll down to switch the toggle just to see what nucleotide mutations were on a branch! We now show both.
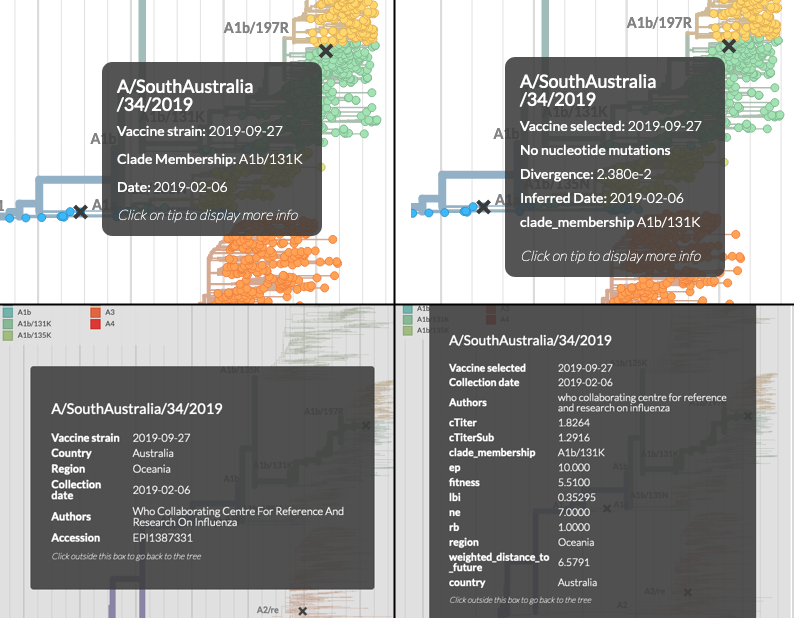 Auspice v1 (left) & v2 (right). v2 shows more information on both tree hover (upper panel) & when clicking on tips (lower panel).
Auspice v1 (left) & v2 (right). v2 shows more information on both tree hover (upper panel) & when clicking on tips (lower panel).
# Display of second trees
Auspice has had the ability to display two trees side-by-side for a while now (and finally documented). If you wanted to, say, compare influenza HA & NA trees, the URL used to look like "flu/seasonal/h3n2/ha:na/2y". This turned out to be problematic when coming up with suitable candidates for potential second-trees, and also made it impossible to compare, for instance, "ha/2y" with "na/3y"
We now use a more verbose syntax to define the display of multiple trees, specifying the entire pathname for both datasets. The above example is now "flu/seasonal/h3n2/ha/2y:flu/seasonal/h3n2/na/2y". Any available datasets can be compared using this URL syntax, even if the result is rather nonsensical. The old syntax will continue to work and will automatically correct to the new syntax (and show you a warning).
P.S. The list of available second trees, which is displayed in the sidebar, is now handed to Auspice by the getAvailable API request.
# Display better dates on the tree axis
Internally, we use decimal dates (e.g. 2012.3 is around the start of may) so that's what we displayed on the tree. It turns out this is pretty hard to interpret when looking at small timespans! We now (a) show dates on the tree's x-axis using months & days, depending on the timespan displayed, and (b) try to use more informative grid spacings. These help with the interpretation of trees over smaller time scales. See PR #804.
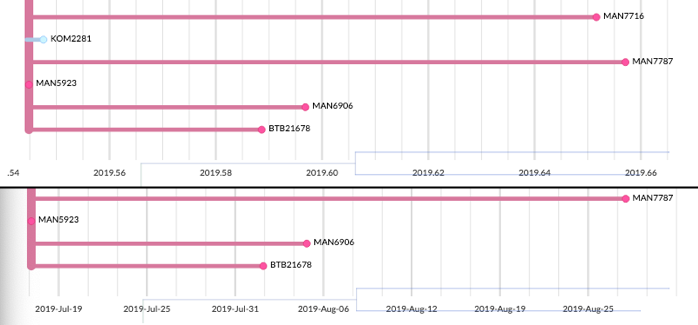 Above: Auspice v1's decimal labels were somewhat hard to interpret. Below: v2 displays calendar dates as appropriate, and uses more intelligent grid spacing.
Above: Auspice v1's decimal labels were somewhat hard to interpret. Below: v2 displays calendar dates as appropriate, and uses more intelligent grid spacing.
# Map "reset zoom" button zooms to include all demes
There's now a button at the top-right of the map which will trigger the map to reset the zoom. See PR #802.
# Consistent colouring of missing data in the tree
If your analysis produces results in - (gaps), X (unknown residue) or N (unknown nucelotide) then we now colour these grey, making it much easier to see when data is missing.
See PR #799.
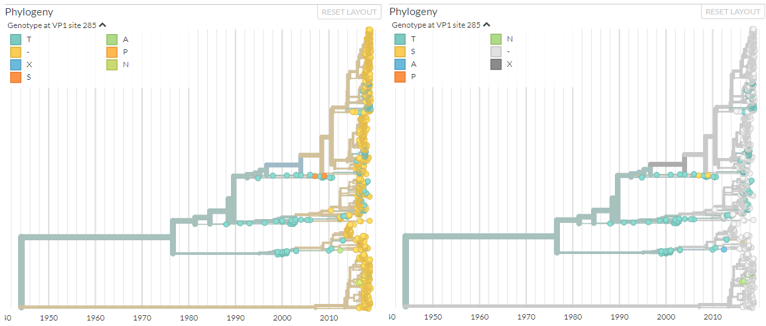 Same analysis, different colour schemes, different interpretation.
Same analysis, different colour schemes, different interpretation.
# Removal of Twitter & Google Analytics
These were a holdover from the early days when nextstrain.org and Auspice were the same thing. We've now removed all calls to Twitter, and made Google Analytics opt in. See requests made from the client for details on exactly what requests are made and how to opt-in to Google Analytics if you desire.
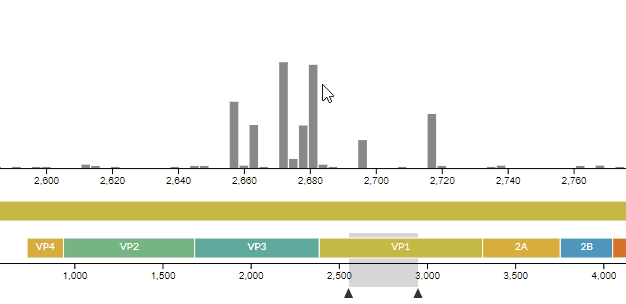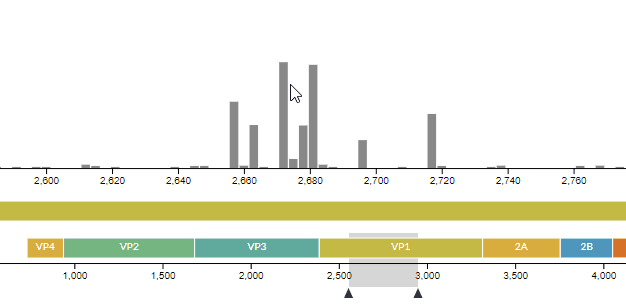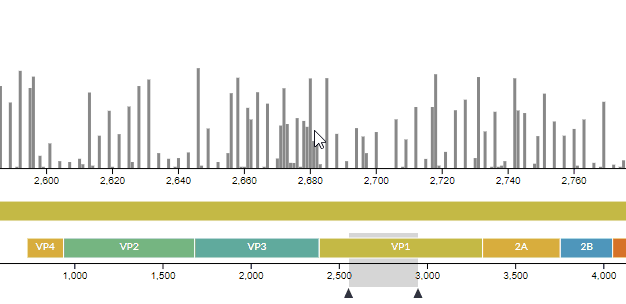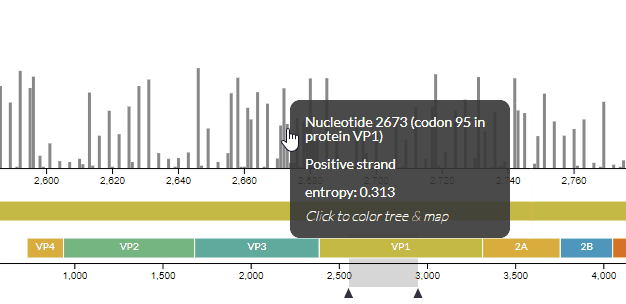
# Improvements in the entropy panel
We improved the usability of the entropy (genomic diversity) panel, as well as fixing a few hidden bugs -- see PR #771. For instance, you can now see which codon a nucleotide codes for (and vice-versa).
# Auspice responds to server redirects for datasets
This allows custom servers (nextstrain.org, for instance!) to smoothly inform Auspice that, e.g., a getDataset request to "/flu" (which doesn't actually exist) should be "/flu/seasonal/h3n2/ha/3y".
See PR #778.
# Importing (server) code from Auspice
Auspice now makes a few helper commands available for those who are writing custom Auspice servers. See these docs for more info.
#
New Auspice subcommand: auspice convert
This is a utility command to convert between dataset formats. Currently, it only converts "Auspice v1" JSONs into "Auspice v2" JSONs, using the same code that is programatically importable.
Right now, auspice view will automatically convert "v1" JSONs into "v2" JSONs, so there's no need to do this yourself.
# Ability to show a "build" source URL in the sidebar
Auspice used to contain some hard-coded logic which was used by nextstrain to display a link to the GitHub repo behind community URLs.
We have now generalised this, and the getAvailable API request can define a buildUrl property for each dataset which auspice will display in the sidebar.
#
auspice view uses a custom Auspice client if present
It's possible to use auspice build to build a custom auspice client.
If this has been done, then running auspice view will serve it -- before you had to run auspice view --customBuild.
This streamlines generating custom auspice bundles and serving them locally.


















