SARS-CoV-2 clade naming strategy for 2022
Cornelius Roemer, Emma B Hodcroft, Richard A Neher, Trevor Bedford
The rapid dominance and increased diversity of the Omicron variant and its constituent sub-lineages has triggered another update of the Nextstrain clade naming guidelines and labels. Once again, we propose a backwards-compatible update, this time to allow more flexible and faster designation of clades as new variants appear and spread.
Our initial strategy of the 'year-letter' naming has been successful in keeping Nextstrain clade names short, clear, and easy to pronounce, and over two years into the pandemic, is accomplishing our original goal of remaining a long-term solution for distinct clade naming where the aim is to have a level of granularity that matches significant differences in biological impact or circulation patterns among viruses Nextstrain clades are coarser than the fine grained Pango lineages.
The updated clade designation rules from last year recognized that reduction in global travel meant that our original clade designation guidelines were not working as intended, and we revised them to allow recognition of spread on regional levels, as well as designation of a variant of concern (VOC) . Through 2021, this labeling worked well, and we designated 21A through 21L (which includes VOCs Delta through Omicron). Our previous updates allowed us to avoid renaming clades, as we had hoped, and worked well in-sync with interest in emerging variants identified initially via Pango lineages.
However, with the dominance and diversification of the Omicron family of variants, we are seeing trends in rising variants that are notable, of public and scientific interest, and genetically distinct, yet do not yet meet previous criteria to be labeled. In order to allow easier reference to such variants at an earlier time, we propose an update to the current guidelines. This will allow designation of a clade if it shows consistent >0.05 per day growth in frequency where it is circulating, in addition to reaching >5% regional frequency.
Our revised set of guidelines will therefore now be to designate a clade when any of the following criteria are met:
- A VOC or VOI is recognized by the WHO and given a Greek letter label
- A clade reaches >20% global frequency for 2 or more months
- A clade reaches >30% regional frequency for 2 or more months
- A clade shows consistent >0.05 per day growth in frequency where it's circulating and has reached >5% regional frequency
Additionally, for points 2, 3, and 4, we require the clade to show multiple mutations in S1 or particular mutations of known biological relevance.
As a result of this, we are designating 3 new Nextstrain clades:
22A- a descendant of d 21L, bearing S:69-70del, S:L452R, S:F486V, reversion S:R493Q, N:P151S, ORF1a:141-143del corresponding to Pango lineage BA.422B- a descendant- of 21L, bearing S:69-70del, S:L452R, S:F486V, reversion S:R493Q, M:D3N and ORF6:L61D, corresponding to Pango lineage BA.522C- a descendant of 21L, bearing S:L452Q and S:S704L, corresponding to Pango lineage BA.2.12.1
As all of these variants are part of the Omicron family and the Omicron group is designated a VOC according to WHO, they will retain the parenthetical "(Omicron)" designation. Note that the topology at the base of clade 21L that contains Pango lineages BA.2, BA.4 and BA.5 is not completely certain. BA.2, BA.4 and BA.5 are designated as sister lineages. Here, we’ve placed clade 21L along with 22A and 22B within 21L to be robust to noise in phylogenetic reconstruction.
You can view updated clade definitions and detailed rationale behind elevating the new clades in this GitHub PR, and the resulting Nextstrain outputs can be seen at nextstrain.org/ncov/.
An updated version of our clade schema diagram is now:
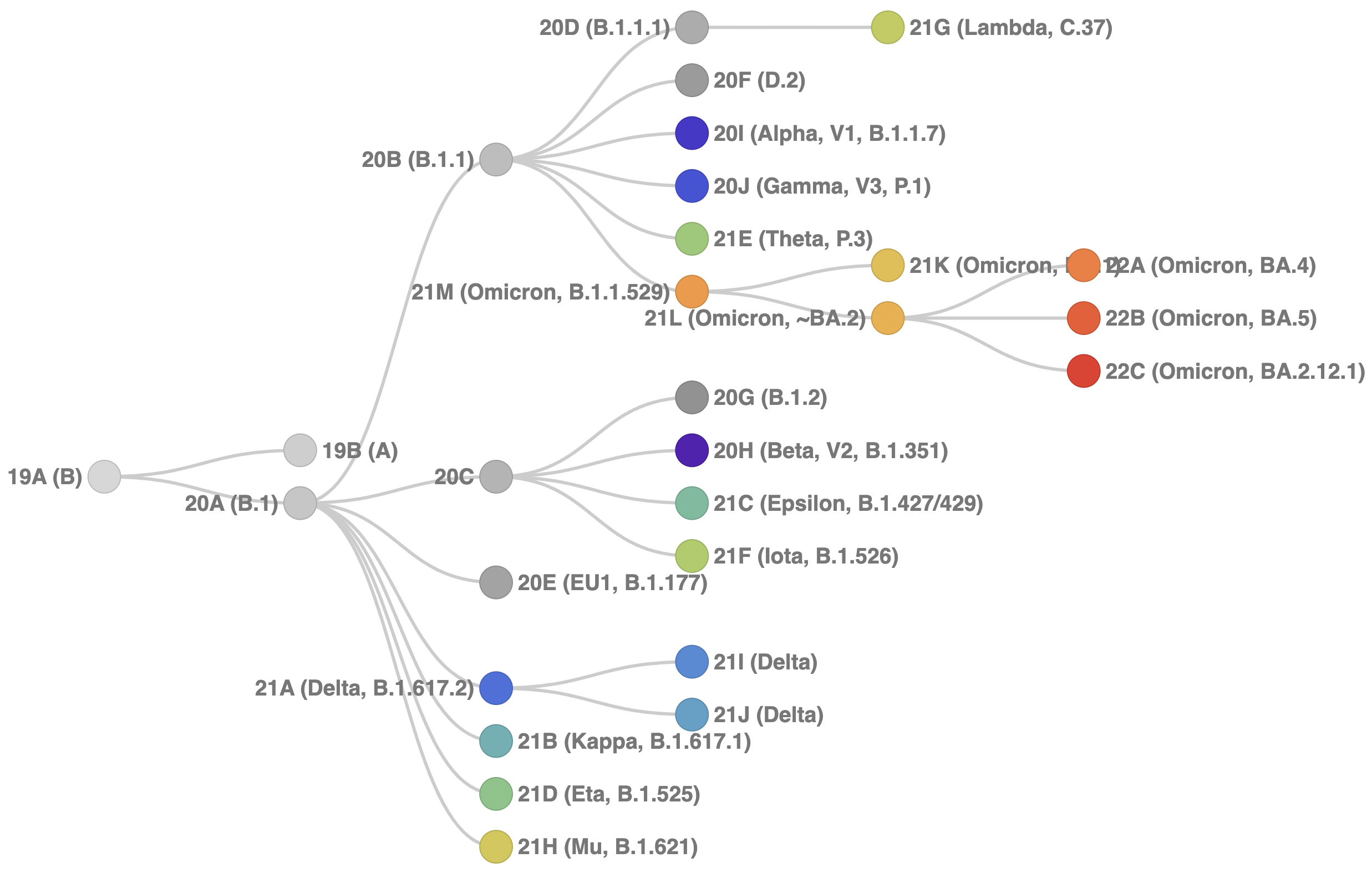 Fig 1. Schematic showing hierarchical relationships among clades. The code behind this diagram is available here.
Fig 1. Schematic showing hierarchical relationships among clades. The code behind this diagram is available here.


















