Extension of SARS-CoV-2 data processing to incorporate Open Data through GenBank
Trevor Bedford, James Hadfield, Emma Hodcroft, John Huddleston, Richard Neher, Thomas Sibley
Progress towards large-scale real-time genomic surveillance of SARS-CoV-2 has been remarkable with over 2 million viral genomes shared to GISAID from all over the world since Jan 2020. This has allowed detailed tracking of the emergence and spread of variants of concern and is an essential pandemic response activity. In an unprecedented collective effort, these data were shared by hundreds of laboratories from all over the world, often within days or weeks of sample collection, and curated by GISAID. Such early sharing of unpublished data requires certain guarantees that submitters get credit for their contributions and a chance to publish on their data. Sequences submitted to the GISAID EpiCoV database are shared under Terms of Use that restrict resharing of sequence data or metadata.
With Nextstrain, we’ve done our best to respect these Terms of Use and we encourage data generators to share data to GISAID, which allows for comprehensive analyses. With Augur, Auspice and our ncov workflow, we have provided academic and public health bioinformaticians around the world with open-source tools to analyze these GISAID data. But while guarantees such as those provided by GISAID’s Terms of Use are important, these restrictions also constrict and constrain data reuse, thereby adding friction to genomic analyses conducted by researchers and public health agencies.
Our goal with Nextstrain is to harness the scientific and public health potential of pathogen genome data and we’ve built our software to be agnostic to the source of data. Genomic surveillance efforts particularly in the US, UK, Germany, Switzerland, Australia, Bangladesh, Chile, Egypt, Ghana, India, Kenya, and Peru have been sharing SARS-CoV-2 genomic sequence data to INSDC databases, which mirror data between GenBank, ENA and DDBJ. Data shared to GenBank and other INSDC databases is shared under Open Data principles. "Open Data is data that can be freely used, re-used and redistributed by anyone - subject only, at most, to the requirement to attribute and sharealike". This allows us at Nextstrain to share sequence data and metadata from GenBank, as well as intermediate files, in an open fashion and help to facilitate data use by academic and public health bioinformaticians.
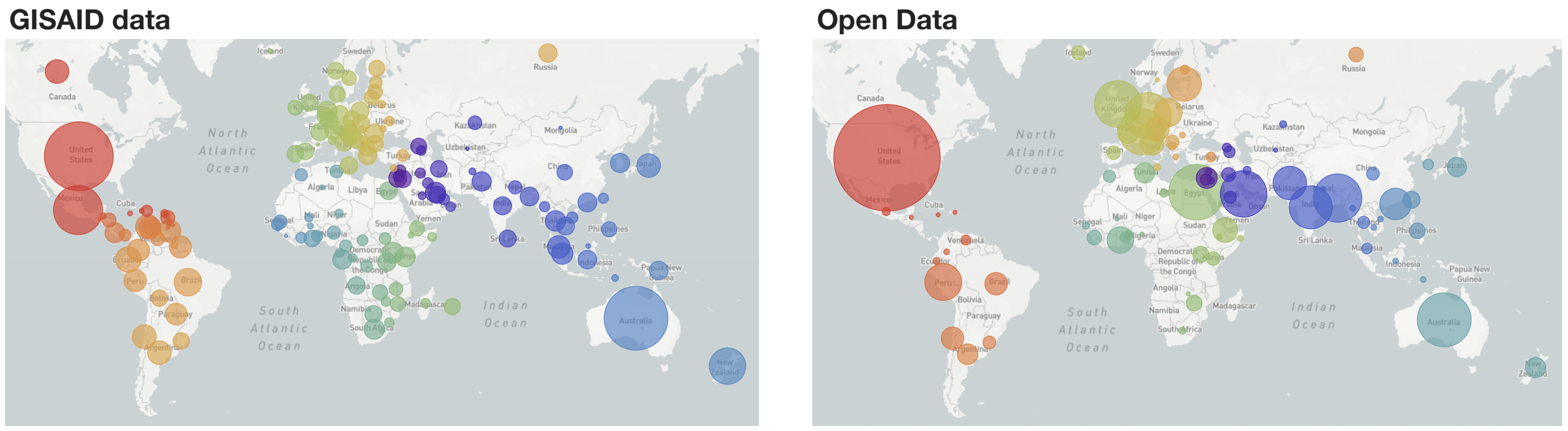 Fig 1. Geographic distribution of samples included in "global" build with GISAID data and Open Data
Fig 1. Geographic distribution of samples included in "global" build with GISAID data and Open Data
Moving forward, we will be simultaneously curating and keeping continually updated analyses for GISAID data at nextstrain.org/ncov/gisaid as well as Open Data at nextstrain.org/ncov/open, where we are mirroring the pattern of one "global" build alongside six "regional" builds (Fig. 1). Open data is fetched from NCBI GenBank in a completely separate process from data fetched from GISAID. To maximize the utility and visibility of shared open datasets, we also provide preprocessed files that can serve as a starting point for additional analyses, including flat files of aligned sequences and curated metadata. Please see our documentation on how to use these preprocessed files. Briefly, this allows Nextstrain build config files to point at URLs like data.nextstrain.org/files/ncov/open/metadata.tsv.gz, providing an automated starting point which lets users avoid having to click through a web interface to provision data. This should allow for more automated rebuilds of public health dashboards. Following Terms of Use, we keep GISAID data secure and only provide access to these intermediate data files for GenBank data.
Please note that although data generators have generously shared data in an open fashion, that does not mean there should be a free license to publish on this data. At Nextstrain we strongly believe in appropriate credit and recognise that "scooping" can make future data generation harder and less appealing to share, and can be particularly problematic for scientists from low and middle income countries. Data generators should be cited where possible and collaborations should be sought in some circumstances. To facilitate such attribution, the open metadata includes links to the original records as well as author information where available. Please try to avoid scooping someone else's work, be mindful of what plans others might have, and reach out if uncertain.
The degree of sequencing and sequence sharing during the pandemic has been remarkable. We thank data submitters from all over the world for generously sharing sequencing data, alongside GISAID and NCBI for providing platforms to easily share this sequence data.


















